1. Feedforward Neural Network Models#
In this tutorial, we will learn how to use a neural network model to predict the energy of points on the Müller-Brown potential energy surface.
1.1. Defining the Müller-Brown Potential Energy Function#
For the definition of Müller-Brown potential, see here.
\(v(x,y) = \sum_{k=0}^3 A_k \mathrm{exp}\left[ a_k (x - x_k^0)^2 + b_k (x - x_k^0) (y - y_k^0) + c_k (y - y_k^0)^2 \right] \)
from math import exp, pow
def mueller_brown_potential(x, y):
A = [-200, -100, -170, 15]
a = [-1, -1, -6.5, 0.7]
b = [0, 0, 11, 0.6]
c = [-10, -10, -6.5, 0.7]
x0 = [1, 0, -0.5, -1.0]
y0 = [0, 0.5, 1.5, 1]
z = 0
for k in range(4):
# Scale the function by 0.1 to make plotting easier
z += 0.1 * A[k] * exp(a[k] * pow(x-x0[k], 2) + b[k] * (x-x0[k]) * (y-y0[k]) + c[k] * pow(y-y0[k], 2))
return z
1.1.1. Generate Training Data#
First, we need to generate data to train the neural network.
import numpy as np
# generate x and y on a grid
x_range = np.arange(-1.8, 1.4, 0.1, dtype=np.float32)
y_range = np.arange(-0.4, 2.4, 0.1, dtype=np.float32)
X, Y = np.meshgrid(x_range, y_range)
# compute the potential energy at each point on the grid
mueller_brown_potential_vectorized = np.vectorize(mueller_brown_potential, otypes=[np.float32])
Z = mueller_brown_potential_vectorized(X, Y)
# keep only low-energy points for training
train_mask = Z < 10
X_train, Y_train, Z_train = X[train_mask], Y[train_mask], Z[train_mask]
print(f"Z_min: {np.min(Z)}, Z_max: {np.max(Z)}")
print(f"Size of (future) training set: {len(Z_train)}")
Z_min: -14.599802017211914, Z_max: 1194.4622802734375
Size of (future) training set: 696
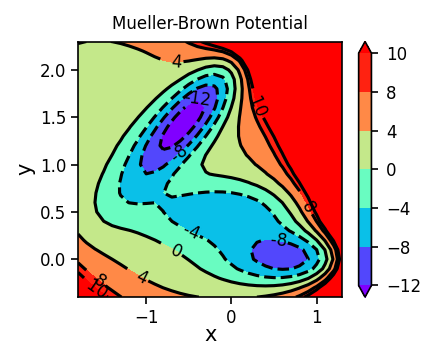
1.1.2. Visualizing Training Data: 3-D Projection Surface#
We will now create a 3-D plot of our training data. We are going to use the plotly library to create an interactive plot. We only plot the part of the potential energy surface that is below 15 (arbitrary unit). For the actual training, only the part that is below 10 (arbitrary unit) will be used.
import plotly.graph_objects as go
fig = go.Figure(data=[go.Surface(z=Z, x=X, y=Y, colorscale='rainbow', cmin=-15, cmax=9)])
fig.update_traces(contours_z=dict(show=True, project_z=True))
fig.update_layout(title='Mueller-Brown Potential', width=500, height=500,
scene = dict(
zaxis = dict(dtick=3, range=[-15, 15]),
camera_eye = dict(x=-1.2, y=-1.2, z=1.2)))
1.1.3. Visualizing Training Data: Contour Surface#
Now we will create a contour surface plot of our training data. Since we will plot similar contour surface plots later, we will define a function to do this.
import matplotlib.pyplot as plt
def plot_contour_map(X, Y, Z, ax, title, colorscale='rainbow', levels=None):
if levels is None:
levels = [-12, -8, -4, 0, 4, 8, 10]
ct = ax.contour(X, Y, Z, levels, colors='k')
ax.clabel(ct, inline=True, fmt='%3.0f', fontsize=8)
ct = ax.contourf(X, Y, Z, levels, cmap=colorscale, extend='both', vmin=levels[0], vmax=levels[-1])
ax.set_xlabel("x", labelpad=-0.75)
ax.set_ylabel("y", labelpad=2.5)
ax.tick_params(axis='both', pad=2, labelsize=8)
cbar = plt.colorbar(ct)
cbar.ax.tick_params(labelsize=8)
ax.set_title(title, fontsize=8)
fig, ax = plt.subplots(figsize=(3, 2.5), dpi=150)
plot_contour_map(X, Y, Z, ax=ax, title='Mueller-Brown Potential')
fig.tight_layout()
1.2. Defining the Neural Network Class#
1.2.1. A Basic Neural Network#
Below is a schematic of a neural network. Inputs (\(x\), \(y\)) are given weights (\(w\)). For each neuron in the hidden layer a bias (\(b\)) is added to the weighted inputs to generate a value (\(v\)) for the activation function (tanh). The activation function decides the activity (\(v'\)) of each neuron in the hidden layer. Weights (\(w'\)), biases (\(b'\)), and the activation function on the neuron of the output layer produces the prediction (\(V^{pred}\)).
Here we define our neural network as a Python class. A function (\(\it{train\_loop}\)) is used to loop through our training data.
import torch
import torch.nn as nn
import torch.nn.functional as F
# Set default data type to float32
torch.set_default_dtype(torch.float32)
class NeuralNetwork(nn.Module):
def __init__(self, n_hidden=20):
"""
Args:
n_hidden (int): Number of neurons in the hidden layer.
"""
super().__init__()
self.linear = nn.Sequential(
nn.Linear(2, n_hidden), # Linear function taking 2 inputs and outputs data for n_hidden neurons.
nn.Tanh(),
nn.Linear(n_hidden, 1) # Linear function taking data from n_hidden neurons and producing one output value.
)
def forward(self, x):
return self.linear(x)
def train_loop(dataloader, model, loss_fn, optimizer):
model.train() # Set model to training mode.
num_batches = len(dataloader)
train_loss = 0
for X, y in dataloader:
# Compute prediction and loss
pred = model(X)
loss = loss_fn(pred.squeeze(), y)
# Backpropagation - using the loss function gradients to update the weights and biases.
optimizer.zero_grad() # Zero out the gradients from the previous iteration to replace them.
loss.backward() # Update the gradients of the loss function.
optimizer.step() # Uses step() to optimize the weights and biases with the updated gradients.
with torch.no_grad():
train_loss += loss.item()
train_loss /= num_batches
return train_loss
1.3. Loading PyTorch and Training Data#
After installing and importing pytorch, we will save our training data as a tensor data set.
from torch.utils.data import TensorDataset, DataLoader
# turn NumPy arrays into PyTorch tensors
X_tensor = torch.from_numpy(np.column_stack((X_train, Y_train)))
Y_tensor = torch.from_numpy(Z_train)
dataset = TensorDataset(X_tensor, Y_tensor)
print(f"Size of training set: {len(dataset)}")
Size of training set: 696
1.4. Training the Model#
Now we can train the neural network model. We will finish our training when the desired number of epochs has been reached. We will also define some terms used for training below.
Epochs - Number of forward/backward passes through the entire neural network.
Learning Rate - Determines the step size as we try to minimize the loss function. A faster learning rate would have a larger step size.
Stochastic Gradient Descent (SGD) - The algorithm used for minimizing the loss function.
Note: In this example, there are 696 training points broken into 87 batches of batch size 8.
# Set random seed for reproducibility
torch.manual_seed(314)
# Hyperparameters
batch_size = 32
epochs = 1000
learning_rate = 1e-2
n_hidden = 20
train_loader = DataLoader(dataset, batch_size=batch_size, shuffle=True)
model = NeuralNetwork(n_hidden)
loss_fn = nn.MSELoss()
optimizer = torch.optim.SGD(model.parameters(), lr=learning_rate)
print("Training Errors:")
for epoch in range(epochs):
train_loss = train_loop(train_loader, model, loss_fn, optimizer)
if (epoch + 1) % 100 == 0:
print(f"epoch: {epoch:>3d} loss: {train_loss:>6.3f}")
print("Done with Training!")
Training Errors:
epoch: 99 loss: 8.242
epoch: 199 loss: 2.019
epoch: 299 loss: 1.576
epoch: 399 loss: 1.281
epoch: 499 loss: 1.142
epoch: 599 loss: 0.992
epoch: 699 loss: 0.871
epoch: 799 loss: 0.842
epoch: 899 loss: 0.785
epoch: 999 loss: 0.745
Done with Training!
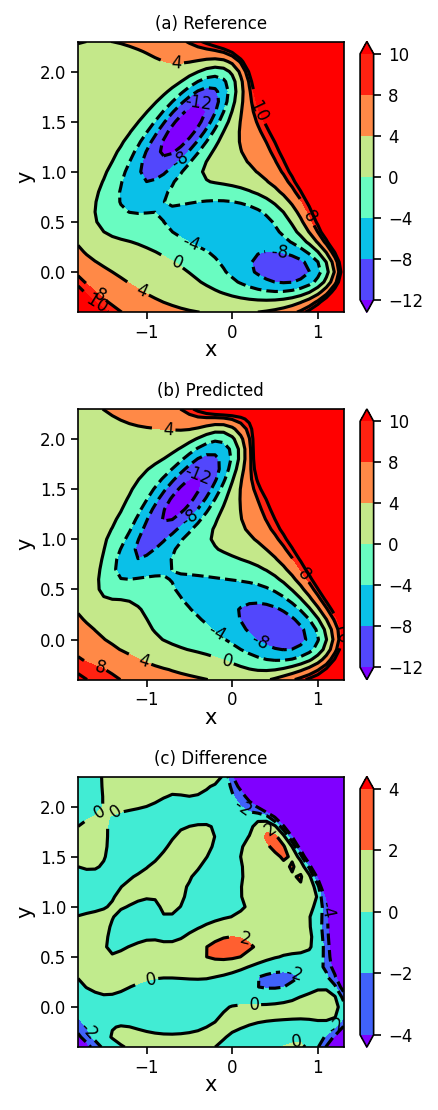
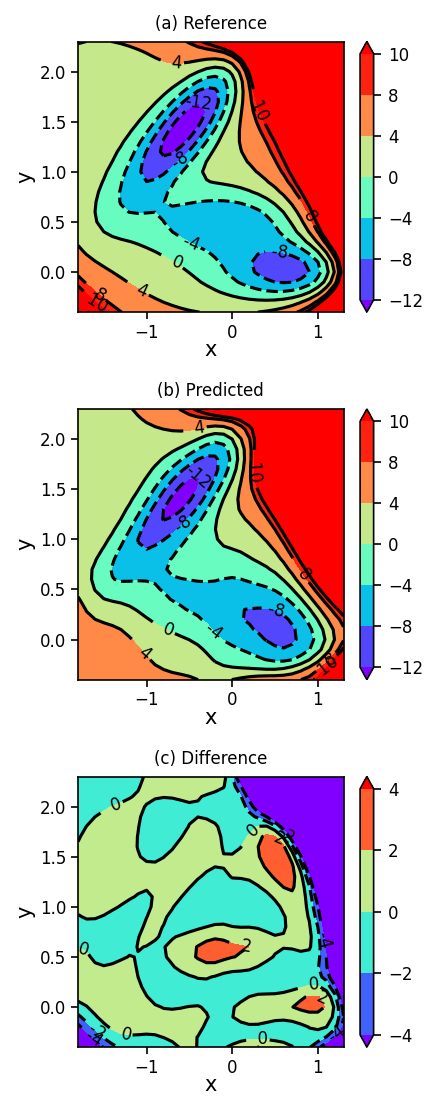
1.5. Plotting Reference, Predicted, and Difference Surfaces#
Finally, we will plot the Müller-Brown potential energy surface using the analytical function (reference), using the neural network (predicted), and we will show the difference between the predicted and reference surfaces.
model.eval() # Set model to evaluation mode
Z_pred = model(torch.from_numpy(np.column_stack((X.flatten(), Y.flatten())))).detach().numpy().reshape(Z.shape)
Z_diff = Z_pred - Z
fig, ax = plt.subplots(3, figsize=(3, 7.5), dpi=150)
plot_contour_map(X, Y, Z, ax=ax[0], title='(a) Reference')
plot_contour_map(X, Y, Z_pred, ax=ax[1], title='(b) Predicted')
plot_contour_map(X, Y, Z_diff, ax=ax[2], title='(c) Difference', levels=[-4, -2, 0, 2, 4])
fig.tight_layout()
1.6. Taking a Look at the NN parameters#
In order to take a closer look at the neural network parameters, we will plug them into the linear functions.
print("model:", model)
for name, param in model.named_parameters():
if name == 'linear.0.weight': weights0 = param.data.detach().numpy()
elif name == 'linear.0.bias': bias0 = param.data.detach().numpy()
elif name == 'linear.2.weight': weights2 = param.data.detach().numpy()
elif name == 'linear.2.bias': bias2 = param.data.detach().numpy()
xy0 = torch.tensor([-0.5, 1.5], dtype=torch.float32)
z0 = model(xy0)
#first linear function
v1 = np.zeros(n_hidden)
for i in range(n_hidden):
v1[i] += weights0[i, 0] * xy0[0] + weights0[i, 1] * xy0[1] + bias0[i]
#activation function
v2 = np.zeros(n_hidden)
for i in range(n_hidden):
v2[i] = np.tanh(v1[i])
#second linear function
z_pred = 0.0
for i in range(n_hidden):
z_pred += weights2[0,i] * v2[i]
z_pred += bias2[0]
print("z0:", z0, "z_pred:", z_pred)
model: NeuralNetwork(
(linear): Sequential(
(0): Linear(in_features=2, out_features=20, bias=True)
(1): Tanh()
(2): Linear(in_features=20, out_features=1, bias=True)
)
)
z0: tensor([-13.0163], grad_fn=<ViewBackward0>) z_pred: -13.016342414391445
1.7. A More Automated/Refined Implementation#
1.7.1. A PyTorch-Lightning Implementation#
Below is a more professional implementation of the neural network that saves epoch infromation in the logs_csv/ directory.
%pip install lightning --quiet
import lightning.pytorch as pl
from lightning.pytorch.loggers import CSVLogger
Note: you may need to restart the kernel to use updated packages.
class NeuralNetworkLightning(pl.LightningModule):
def __init__(self, model, learning_rate):
super().__init__()
self.model = model
self.learning_rate = learning_rate
def forward(self, x):
return self.model(x)
def training_step(self, batch, batch_idx):
x, y = batch
y_pred = self.model(x)
loss = F.mse_loss(y_pred.squeeze(), y)
self.log('train_loss', loss)
return loss
def configure_optimizers(self):
# Using the Adam optimzation algorithm instead of SGD.
optimizer = torch.optim.Adam(self.parameters(), lr=self.learning_rate)
return [optimizer]
%%capture
# Set random seed for reproducibility
pl.seed_everything(314, workers=True);
# Hyperparameters
batch_size = 32
epochs = 1000
learning_rate = 1e-2
n_hidden = 20
train_loader = DataLoader(dataset, batch_size=batch_size, shuffle=True)
csv_logger = CSVLogger('logs_csv', version=0)
trainer = pl.Trainer(max_epochs=epochs, logger=csv_logger, deterministic=True)
model = NeuralNetworkLightning(NeuralNetwork(n_hidden), learning_rate)
trainer.fit(model, train_loader)
Seed set to 314
GPU available: False, used: False
TPU available: False, using: 0 TPU cores
IPU available: False, using: 0 IPUs
HPU available: False, using: 0 HPUs
| Name | Type | Params
----------------------------------------
0 | model | NeuralNetwork | 81
----------------------------------------
81 Trainable params
0 Non-trainable params
81 Total params
0.000 Total estimated model params size (MB)
`Trainer.fit` stopped: `max_epochs=1000` reached.
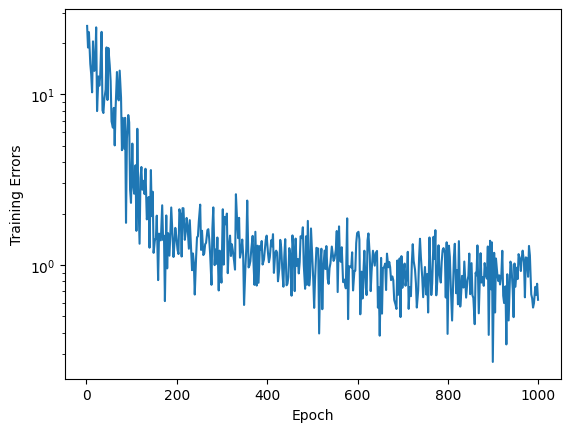
1.7.2. Plotting Training Error#
Now we can plot the training error as the number of epochs increases.
import pandas as pd
loss = pd.read_csv("logs_csv/lightning_logs/version_0/metrics.csv")
fig, ax = plt.subplots()
ax.semilogy(loss["epoch"], loss["train_loss"])
ax.set_xlabel("Epoch")
ax.set_ylabel("Training Errors")
Text(0, 0.5, 'Training Errors')
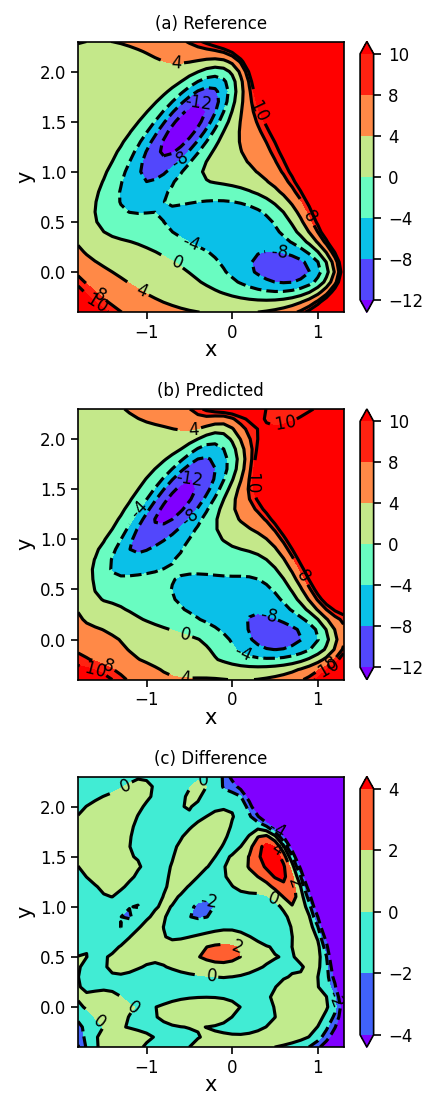
1.7.3. Plotting Reference, Predicted, and Difference Surfaces#
Again, we plot the reference, predicted, and difference surfaces using the more refined neural network implementation.
model.eval() # Set model to evaluation mode
Z_pred = model(torch.from_numpy(np.column_stack((X.flatten(), Y.flatten())))).detach().numpy().reshape(Z.shape)
Z_diff = Z_pred - Z
fig, ax = plt.subplots(3, figsize=(3, 7.5), dpi=150)
plot_contour_map(X, Y, Z, ax=ax[0], title='(a) Reference')
plot_contour_map(X, Y, Z_pred, ax=ax[1], title='(b) Predicted')
plot_contour_map(X, Y, Z_diff, ax=ax[2], title='(c) Difference', levels=[-4, -2, 0, 2, 4])
fig.tight_layout()
1.8. Train with Energy and Gradient#
def mueller_brown_potential_gradient(x, y):
A = [-200, -100, -170, 15]
a = [-1, -1, -6.5, 0.7]
b = [0, 0, 11, 0.6]
c = [-10, -10, -6.5, 0.7]
x0 = [1, 0, -0.5, -1.0]
y0 = [0, 0.5, 1.5, 1]
dx = 0
dy = 0
for k in range(4):
dx += 0.1 * A[k] * exp(a[k] * pow(x-x0[k], 2) + b[k] * (x-x0[k]) * (y-y0[k]) + c[k] * pow(y-y0[k], 2)) * (a[k] * 2 *(x-x0[k]) + b[k] * (y-y0[k]))
dy += 0.1 * A[k] * exp(a[k] * pow(x-x0[k], 2) + b[k] * (x-x0[k]) * (y-y0[k]) + c[k] * pow(y-y0[k], 2)) * (b[k] * (x-x0[k])+ c[k] * 2 * (y-y0[k]))
return dx, dy
# compute the potential energy gradient at each point on the grid
mueller_brown_potential_gradient_vectorized = np.vectorize(mueller_brown_potential_gradient, otypes=[np.float32, np.float32])
dX, dY = mueller_brown_potential_gradient_vectorized(X, Y)
# keep only low-energy points for training
dX_train, dY_train = dX[train_mask], dY[train_mask]
dX_tensor = torch.from_numpy(np.column_stack((dX_train, dY_train)))
dataset = TensorDataset(X_tensor, Y_tensor, dX_tensor)
def train_loop_with_gradient(dataloader, model, loss_fn, optimizer):
model.train() # Set model to training mode.
num_batches = len(dataloader)
train_loss = 0
train_loss_energy = 0
train_loss_gradient = 0
for X, y, dX in dataloader:
# Compute prediction and loss
y_pred = model(X)
dX_pred = torch.autograd.grad(y_pred, X, grad_outputs=torch.ones_like(y_pred), create_graph=True)[0]
loss_energy = loss_fn(y_pred.squeeze(), y)
loss_gradient = loss_fn(dX_pred.squeeze(), dX)
loss = loss_energy + loss_gradient
# Backpropagation - using the loss function gradients to update the weights and biases.
optimizer.zero_grad() # Zero out the gradients from the previous iteration to replace them.
loss.backward() # Update the gradients of the loss function.
optimizer.step() # Uses step() to optimize the weights and biases with the updated gradients.
with torch.no_grad():
train_loss += loss.item()
train_loss_energy += loss_energy.item()
train_loss_gradient += loss_gradient.item()
train_loss /= num_batches
train_loss_energy /= num_batches
train_loss_gradient /= num_batches
return train_loss, train_loss_energy, train_loss_gradient
# Set random seed for reproducibility
torch.manual_seed(314)
# Hyperparameters
batch_size = 32
epochs = 1000
learning_rate = 1e-3
n_hidden = 20
train_loader = DataLoader(dataset, batch_size=batch_size, shuffle=True)
model = NeuralNetwork(n_hidden)
loss_fn = nn.MSELoss()
optimizer = torch.optim.SGD(model.parameters(), lr=learning_rate)
print("Training errors:")
for epoch in range(epochs):
X_tensor.requires_grad_(True)
train_loss, train_loss_energy, train_loss_gradient = train_loop_with_gradient(train_loader, model, loss_fn, optimizer)
if (epoch + 1) % 100 == 0:
print(f"epoch: {epoch:>3d} loss: {train_loss:>6.3f} energy loss: {train_loss_energy:>6.3f} gradient loss: {train_loss_gradient:>6.3f}")
print("Done with Training!")
Training errors:
epoch: 99 loss: 155.851 energy loss: 10.925 gradient loss: 144.926
epoch: 199 loss: 75.487 energy loss: 4.643 gradient loss: 70.844
epoch: 299 loss: 31.353 energy loss: 1.031 gradient loss: 30.322
epoch: 399 loss: 27.340 energy loss: 0.847 gradient loss: 26.493
epoch: 499 loss: 25.616 energy loss: 0.818 gradient loss: 24.797
epoch: 599 loss: 24.661 energy loss: 0.823 gradient loss: 23.838
epoch: 699 loss: 23.933 energy loss: 0.844 gradient loss: 23.089
epoch: 799 loss: 23.246 energy loss: 0.827 gradient loss: 22.418
epoch: 899 loss: 22.167 energy loss: 0.756 gradient loss: 21.412
epoch: 999 loss: 21.067 energy loss: 0.692 gradient loss: 20.375
Done with Training!
model.eval() # Set model to evaluation mode
Z_pred = model(torch.from_numpy(np.column_stack((X.flatten(), Y.flatten())))).detach().numpy().reshape(Z.shape)
Z_diff = Z_pred - Z
fig, ax = plt.subplots(3, figsize=(3, 7.5), dpi=150)
plot_contour_map(X, Y, Z, ax=ax[0], title='(a) Reference')
plot_contour_map(X, Y, Z_pred, ax=ax[1], title='(b) Predicted')
plot_contour_map(X, Y, Z_diff, ax=ax[2], title='(c) Difference', levels=[-4, -2, 0, 2, 4])
fig.tight_layout()